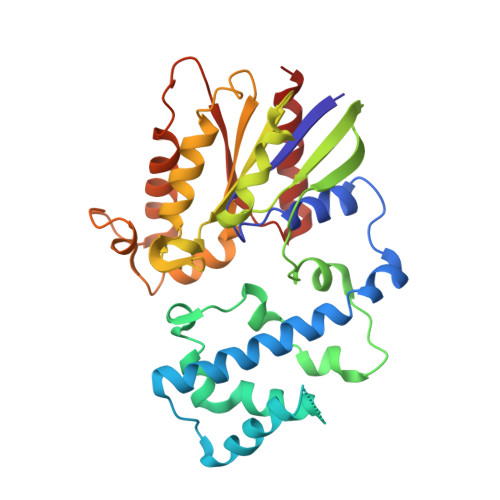
Minimal Determinants for Binding Activated Galpha from the Structure of a Galpha(i1)-Peptide Dimer.
Johnston, C.A., Lobanova, E.S., Shavkunov, A.S., Low, J., Ramer, J.K., Blaesius, R., Fredericks, Z., Willard, F.S., Kuhlman, B., Arshavsky, V.Y., Siderovski, D.P.(2006) Biochemistry 45: 11390-11400
- PubMed: 16981699
- DOI: https://doi.org/10.1021/bi0613832
- Primary Citation of Related Structures:
2G83 - PubMed Abstract:
G-proteins cycle between an inactive GDP-bound state and an active GTP-bound state, serving as molecular switches that coordinate cellular signaling. We recently used phage display to identify a series of peptides that bind G alpha subunits in a nucleotide-dependent manner [Johnston, C. A., Willard, F. S., Jezyk, M. R., Fredericks, Z., Bodor, E. T., Jones, M. B., Blaesius, R., Watts, V. J., Harden, T. K., Sondek, J., Ramer, J. K., and Siderovski, D. P. (2005) Structure 13, 1069-1080]. Here we describe the structural features and functions of KB-1753, a peptide that binds selectively to GDP x AlF4(-)- and GTPgammaS-bound states of G alpha(i) subunits. KB-1753 blocks interaction of G alpha(transducin) with its effector, cGMP phosphodiesterase, and inhibits transducin-mediated activation of cGMP degradation. Additionally, KB-1753 interferes with RGS protein binding and resultant GAP activity. A fluorescent KB-1753 variant was found to act as a sensor for activated G alpha in vitro. The crystal structure of KB-1753 bound to G alpha(i1) x GDP x AlF4(-) reveals binding to a conserved hydrophobic groove between switch II and alpha3 helices and, along with supporting biochemical data and previous structural analyses, supports the notion that this is the site of effector interactions for G alpha(i) subunits.
- Department of Pharmacology, University of North Carolina School of Medicine, Chapel Hill, North Carolina 27599-7365, USA.
Organizational Affiliation: