Novel binding mode for negative allosteric NMDA receptor modulators.
Lotti, J.S., Syrenne, J.T., Benton, A.J., Al-Mousawi, A., Cornelison, L.E., Trolinder, C.J., Yi, F., Zhang, Z., Yates-Hansen, C.K., McClelland, L.J., Bosco, J., Rau, A.R., Clausen, R.P., Hansen, K.B.(2026) J Gen Physiol 158
- PubMed: 41335128
- DOI: https://doi.org/10.1085/jgp.202513872
- Primary Citation of Related Structures:
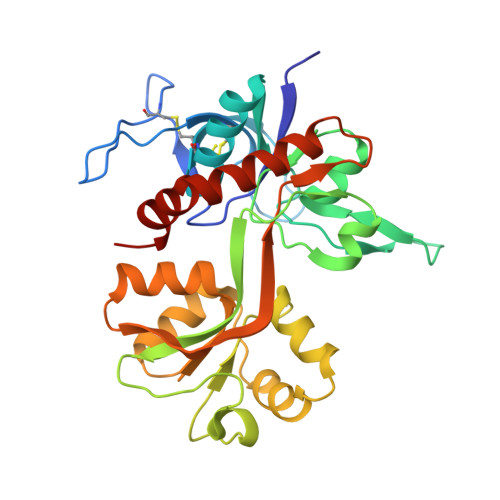
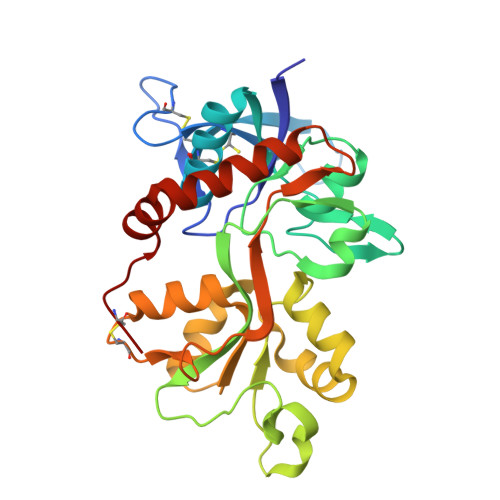
9NYZ - PubMed Abstract:
NMDA-type ionotropic glutamate receptors mediate excitatory neurotransmission and synaptic plasticity, but aberrant signaling by these receptors is also implicated in brain disorders. Here, we present the binding site and the mechanism of action for UCM-101, a novel negative NMDA receptor modulator that produces full inhibition of NMDA receptor-mediated excitatory postsynaptic currents in hippocampal CA pyramidal neurons from juvenile mouse brain slices. UCM-101 has a 59-fold higher binding affinity at GluN1/2A compared with GluN1/2B receptors and inhibits diheteromeric GluN1/2A and triheteromeric GluN1/2A/2B receptors with IC50 values of 110 and 240 nM, respectively, in the presence of 1 µM glycine. The novel binding mode for UCM-101 is revealed in a high-resolution crystal structure of the GluN1/2A agonist binding domain heterodimer. UCM-101 and its analog TCN-213 inhibit NMDA receptors by negatively modulating co-agonist binding to the GluN1 subunit via an allosteric mechanism that is conserved with previously described GluN2A-selective antagonists, TCN-201 and MPX-004. Despite the shared mechanism of action, the structural determinants that mediate subunit selectivity for UCM-101 are distinct from those of TCN-201 and MPX-004. These findings provide detailed insights into the binding site and mechanism of action of a novel NMDA receptor modulator and open new avenues for the development of NMDA receptor ligands with therapeutic potential.
- Center for Structural and Functional Neuroscience, Division of Biological and Biomedical Sciences, University of Montana, Missoula, MT, USA.
Organizational Affiliation: