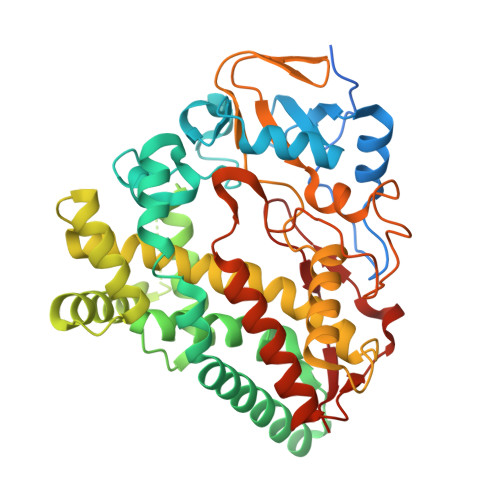
Metabolic engineering of doxorubicin biosynthesis through P450-redox partner optimization and structural analysis of DoxA.
Koroleva, A., Artukka, E., Yamada, K., Newmister, S.A., Harte, R.J., Boesger, H., Londen, M., Sanders, J.N., Tirkkonen, H., Kannisto, M., Kuin, R.C.M., Hulst, M., Wang, R., Leskinen, E., Barillec, M., Niemi, J., van Wezel, G.P., Neefjes, J., Nybo, S.E., Houk, K.N., Sherman, D.H., Kim, R.Q., Metsa-Ketela, M.(2026) Nat Commun
- PubMed: 41639599
- DOI: https://doi.org/10.1038/s41467-026-69194-6
- Primary Citation of Related Structures:
9S7F, 9SI5 - PubMed Abstract:
Doxorubicin, a widely used chemotherapy drug, is produced by Streptomyces peucetius ATCC27952. The biosynthesis relies on the cytochrome P450 monooxygenase DoxA, which catalyzes three consecutive late-stage oxidation steps. However, conversion from daunorubicin to doxorubicin is inefficient, necessitating semi-synthetic industrial manufacturing. Here, we address key limitations in DoxA catalysis. We identify the natural redox partners ferredoxin Fdx4 and ferredoxin reductase FdR3 by transcriptomic analysis. We discovered the vicinal oxygen chelate family protein DnrV to prevent product inhibition by binding doxorubicin. Structural analysis of DoxA and density functional theory (DFT) calculations reveal that inefficient C14 hydroxylation results from the unfavorable anti-conformation of the methyl ketone side chain of daunorubicin. We harness these advances for rational strain engineering, leading to an 180% increase in doxorubicin yields and an improved production profile. This study provides singular insights into enzymatic constraints in anthracycline biosynthesis and facilitates cost-effective manufacturing to meet the growing global demand for doxorubicin.
- Department of Life Technologies, University of Turku, Turku, Finland.
Organizational Affiliation: