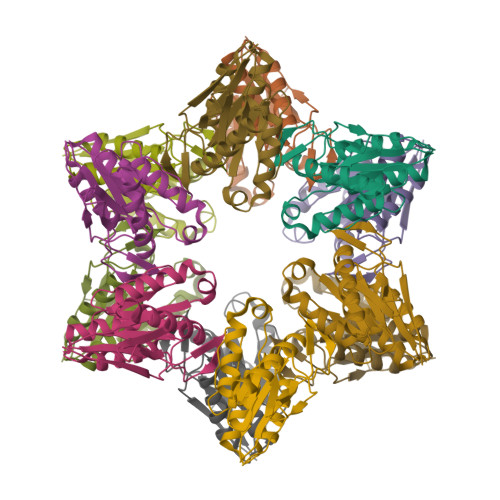
Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Wang, J., Song, J.J., Seong, I.S., Franklin, M.C., Kamtekar, S., Eom, S.H., Chung, C.H.(2001) Structure 9: 1107-1116
- PubMed: 11709174
- DOI: https://doi.org/10.1016/s0969-2126(01)00670-0
- Primary Citation of Related Structures:
1HQY, 1HT1, 1HT2 - PubMed Abstract:
The bacterial heat shock locus ATPase HslU is an AAA(+) protein that has structures known in many nucleotide-free and -bound states. Nucleotide is required for the formation of the biologically active HslU hexameric assembly. The hexameric HslU ATPase binds the dodecameric HslV peptidase and forms an ATP-dependent HslVU protease. We have characterized four distinct HslU conformational states, going sequentially from open to closed: the empty, SO(4), ATP, and ADP states. The nucleotide binds at a cleft formed by an alpha/beta domain and an alpha-helical domain in HslU. The four HslU states differ by a rotation of the alpha-helical domain. This classification leads to a correction of nucleotide identity in one structure and reveals the ATP hydrolysis-dependent structural changes in the HslVU complex, including a ring rotation and a conformational change of the HslU C terminus. This leads to an amended protein unfolding-coupled translocation mechanism. The observed nucleotide-dependent conformational changes in HslU and their governing principles provide a framework for the mechanistic understanding of other AAA(+) proteins.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, 266 Whitney Avenue, New Haven, CT 06520, USA. wang@mail.csb.yale.edu