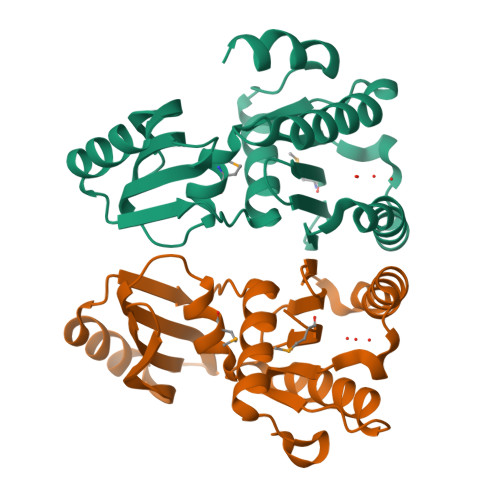
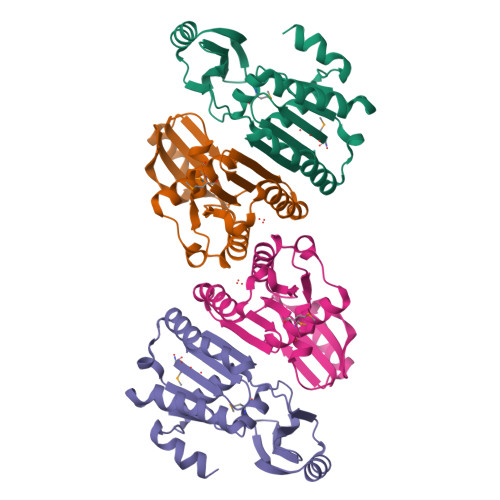
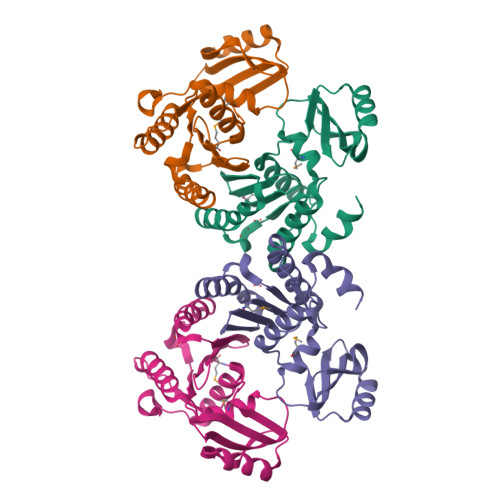
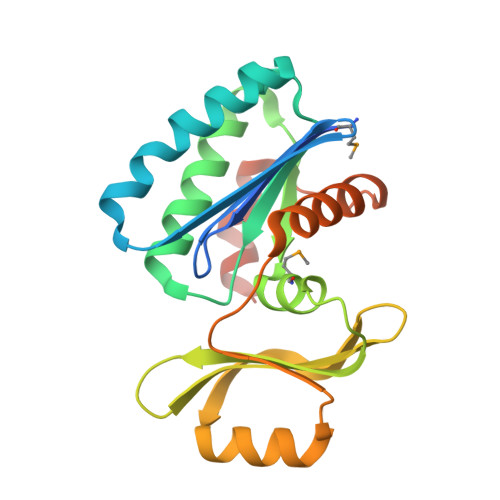
Structure of an essential bacterial protein YeaZ (TM0874) from Thermotoga maritima at 2.5 A resolution.
Xu, Q., McMullan, D., Jaroszewski, L., Krishna, S.S., Elsliger, M.A., Yeh, A.P., Abdubek, P., Astakhova, T., Axelrod, H.L., Carlton, D., Chiu, H.J., Clayton, T., Duan, L., Feuerhelm, J., Grant, J., Han, G.W., Jin, K.K., Klock, H.E., Knuth, M.W., Miller, M.D., Morse, A.T., Nigoghossian, E., Okach, L., Oommachen, S., Paulsen, J., Reyes, R., Rife, C.L., van den Bedem, H., Hodgson, K.O., Wooley, J., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1230-1236
- PubMed: 20944216
- DOI: https://doi.org/10.1107/S1744309109022192
- Primary Citation of Related Structures:
2A6A - PubMed Abstract:
YeaZ is involved in a protein network that is essential for bacteria. The crystal structure of YeaZ from Thermotoga maritima was determined to 2.5 Å resolution. Although this protein belongs to a family of ancient actin-like ATPases, it appears that it has lost the ability to bind ATP since it lacks some key structural features that are important for interaction with ATP. A conserved surface was identified, supporting its role in the formation of protein complexes.
Organizational Affiliation:
Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, Menlo Park, CA, USA.