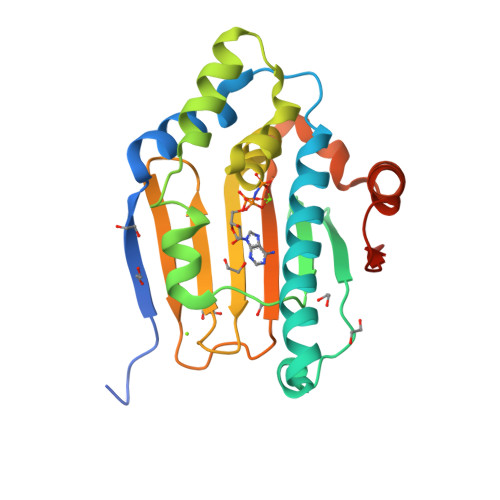
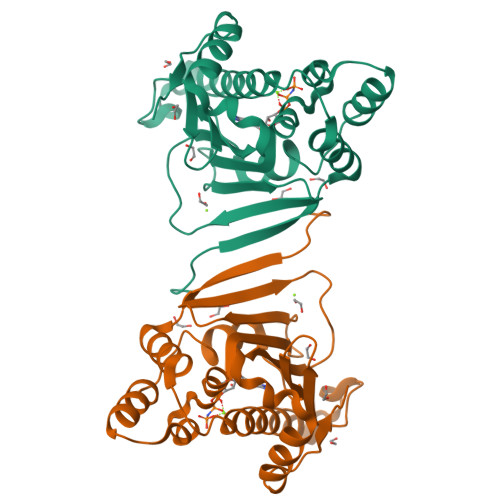
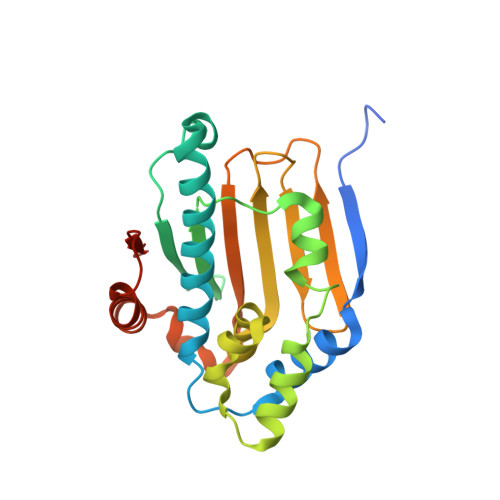
Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213
Wernimont, A.K., Tempel, W., Lin, Y.H., Hutchinson, A., Mackenzie, F., Fairlamb, A., Kozieradzki, I., Cossar, D., Zhao, Y., Schapira, M., Bochkarev, A., Arrowsmith, C.H., Bountra, C., Weigelt, J., Edwards, A.M., Ferguson, M.A.J., Hui, R., Pizarro, J.C., Hills, T.To be published.