N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
Nuti, E., Cantelmo, A.R., Gallo, C., Bruno, A., Bassani, B., Camodeca, C., Tuccinardi, T., Vera, L., Orlandini, E., Nencetti, S., Stura, E.A., Martinelli, A., Dive, V., Albini, A., Rossello, A.(2015) J Med Chem 58: 7224-7240
- PubMed: 26263024
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00367
- Primary Citation of Related Structures:
4WZV, 4XCT - PubMed Abstract:
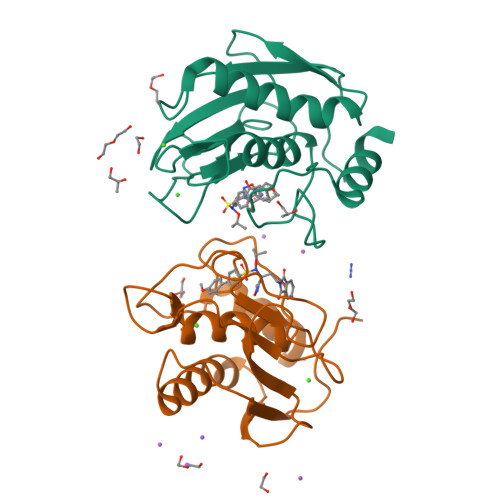
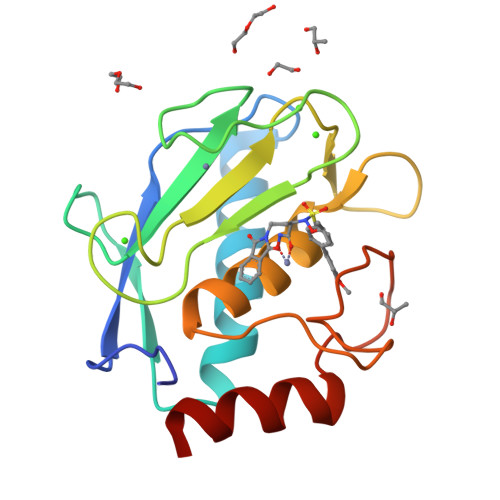
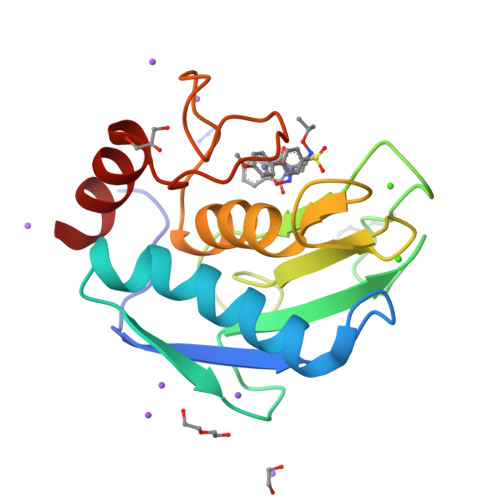
Matrix metalloproteinases (MMPs) have been shown to be involved in tumor-induced angiogenesis. In particular, MMP-2, MMP-9, and MMP-14 have been reported to be crucial for tumor angiogenesis and the formation of metastasis, thus becoming attractive targets in cancer therapy. Here, we report our optimization effort to identify novel N-isopropoxy-arylsulfonamide hydroxamates with improved inhibitory activity toward MMP-2, MMP-9, and MMP-14 with respect to the previously discovered compound 1. A new series of hydroxamates was designed, synthesized, and tested for their antiangiogenic activity using in vitro assays with human umbilical vein endothelial cells (HUVECs). A nanomolar MMP-2, MMP-9, and MMP-14 inhibitor was identified, compound 3, able to potently inhibit angiogenesis in vitro and also in vivo in the matrigel sponge assay in mice. Finally, X-ray crystallographic and docking studies were conducted for compound 3 in order to investigate its binding mode to MMP-9 and MMP-14.
Organizational Affiliation:
Dipartimento di Farmacia, Università di Pisa , via Bonanno 6, 56126 Pisa, Italy.