Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
Monn, J.A., Prieto, L., Taboada, L., Hao, J., Reinhard, M.R., Henry, S.S., Beadle, C.D., Walton, L., Man, T., Rudyk, H., Clark, B., Tupper, D., Baker, S.R., Lamas, C., Montero, C., Marcos, A., Blanco, J., Bures, M., Clawson, D.K., Atwell, S., Lu, F., Wang, J., Russell, M., Heinz, B.A., Wang, X., Carter, J.H., Getman, B.G., Catlow, J.T., Swanson, S., Johnson, B.G., Shaw, D.B., McKinzie, D.L.(2015) J Med Chem 58: 7526-7548
- PubMed: 26313429
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01124
- Primary Citation of Related Structures:
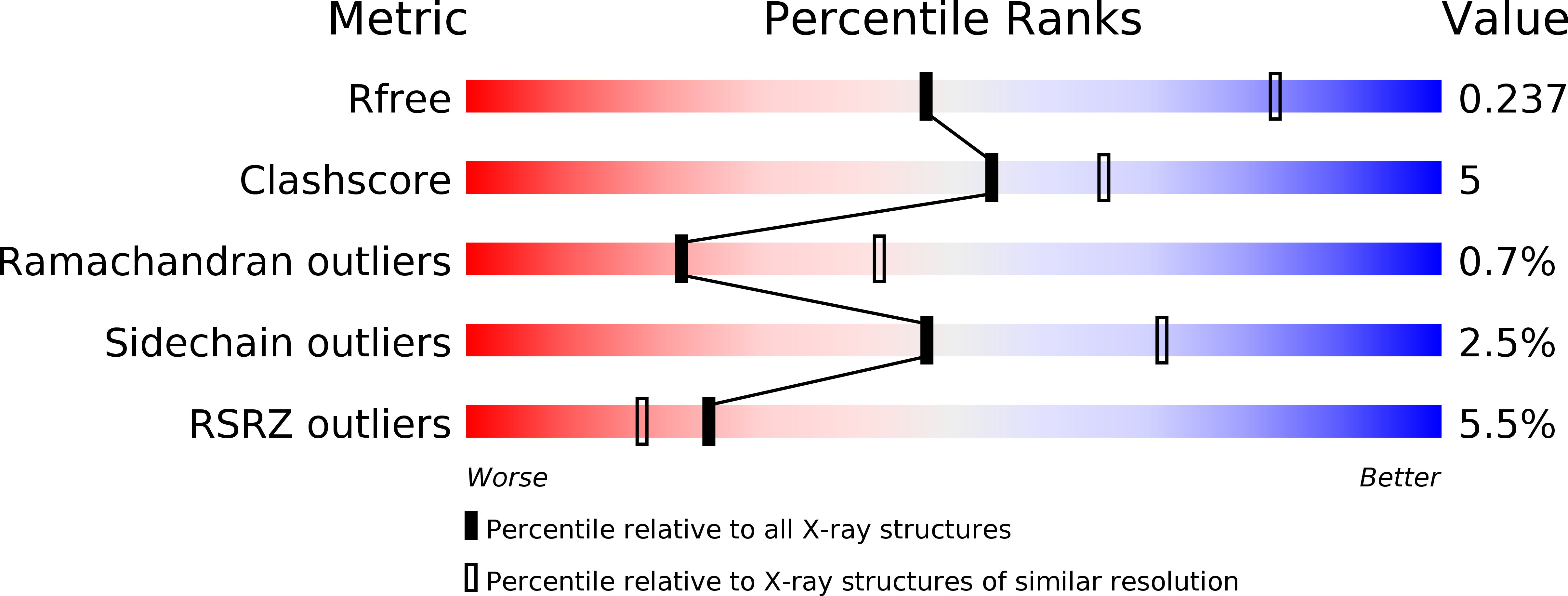
5CNI, 5CNJ, 5CNK, 5CNM - PubMed Abstract:
Identification of orthosteric mGlu(2/3) receptor agonists capable of discriminating between individual mGlu2 and mGlu3 subtypes has been highly challenging owing to the glutamate-site sequence homology between these proteins. Herein we detail the preparation and characterization of a series of molecules related to (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylate 1 (LY354740) bearing C4-thiotriazole substituents. On the basis of second messenger responses in cells expressing other recombinant human mGlu2/3 subtypes, a number of high potency and efficacy mGlu2 receptor agonists exhibiting low potency mGlu3 partial agonist/antagonist activity were identified. From this, (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid 14a (LY2812223) was further characterized. Cocrystallization of 14a with the amino terminal domains of hmGlu2 and hmGlu3 combined with site-directed mutation studies has clarified the underlying molecular basis of this unique pharmacology. Evaluation of 14a in a rat model responsive to mGlu2 receptor activation coupled with a measure of central drug disposition provides evidence that this molecule engages and activates central mGlu2 receptors in vivo.
Organizational Affiliation:
Discovery Chemistry Research and Technologies, ‡Quantitative Biology, §Structural Biology, ∥Drug Disposition and ⊥Neuroscience Research, Eli Lilly and Company , Lilly Corporate Center, Drop 0510, Indianapolis, Indiana 46285, United States.