Structure-Based Rational Design and Evaluation of BET-Aurora Kinase Dual-Inhibitors for Treatment of Cancers.
Lyu, K., Ren, Y., Mou, J., Yang, Y., Pan, Y., Zhang, H., Li, Y., Cao, D., Chen, L., Chen, D., Guo, D., Xiong, B.(2025) J Med Chem 68: 1344-1364
- PubMed: 39844725
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01933
- Primary Citation of Related Structures:
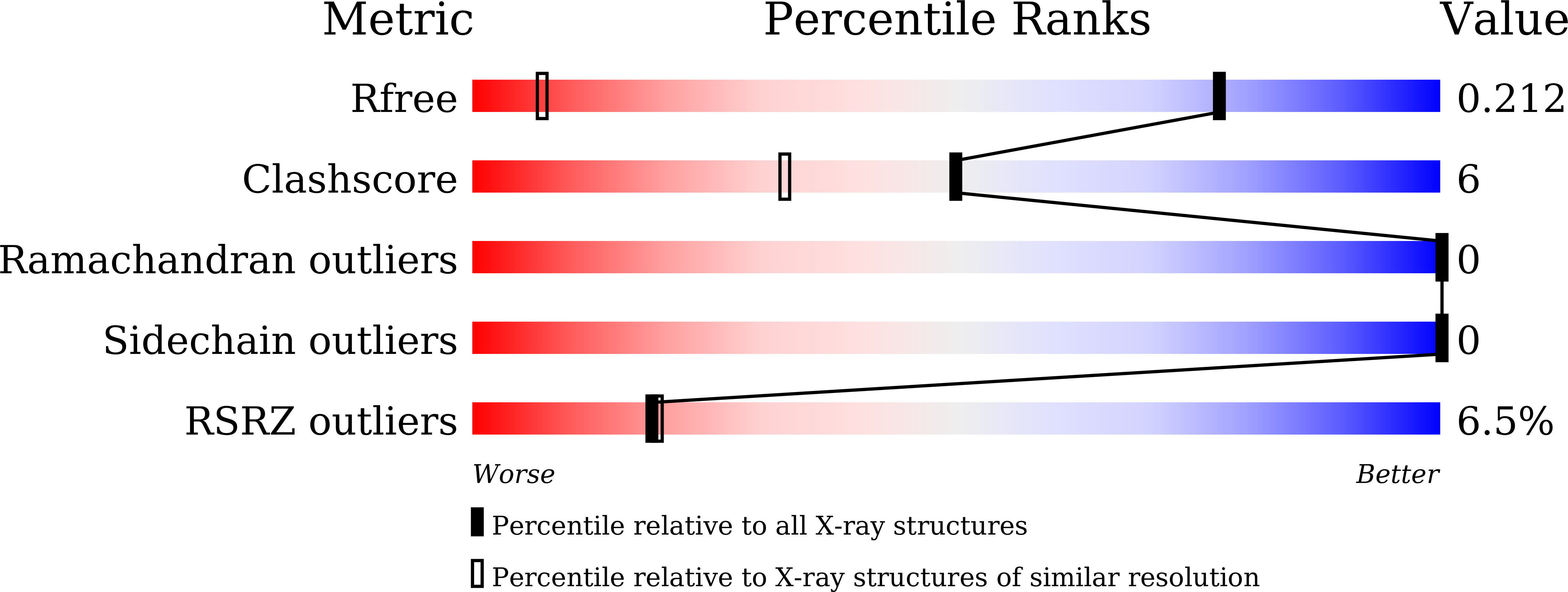
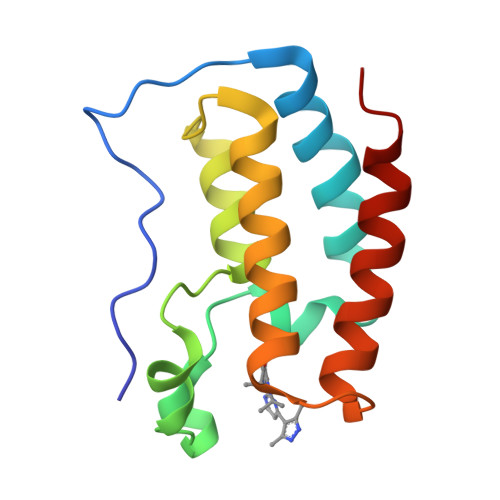
8WQO - PubMed Abstract:
Simultaneous inhibition of the bromodomain and extra-terminal domain and Aurora kinases is a promising anticancer therapeutic strategy. Based on our previous study on BET-kinase dual inhibitors, we employed the molecular docking approach to design novel dual BET-Aurora kinase A inhibitors. Through several rounds of optimization and with the guidance of the solved cocrystal structure of BRD4 bound to inhibitor 27 , we finally obtained a series of highly potent dual BET-Aurora kinase A inhibitors. Compound 38 exhibited strong affinity toward both BRD4 and Aurora kinase A. It also showed good antiproliferative activities on diverse cancer cell lines, good pharmacokinetic profiles, and favorable antitumor efficacy in renal cell cancer and colon cancer xenograft models with TGI of 45.99% and 53.06%, respectively. The development of compound 38 reinforces the concept that a rational design may achieve dual inhibitors targeting specific kinases and bromodomain proteins.
Organizational Affiliation:
Department of Medicinal Chemistry, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, China.