RCSB PDB Help
Search and Browse > Advanced Search
Sequence Similarity Search
Introduction
The Sequence Similarity search helps identify proteins or protein domains that are evolutionarily related by comparing their amino-acid sequences. It is widely used to detect homologs, infer function, and explore relationships across protein families.
Note: Although protein sequences generally determine how a protein folds into its functional 3D shape, some proteins can adopt multiple conformations or even fold into different structures (e.g., prion proteins). To compare molecular shapes or conformational states directly, use a 3D Similarity search instead.
How Sequence Similarity Search Works?
The Sequence Similarity search lets you query sequences from the PDB archive—and optionally available Computed Structure Models (CSMs)—using a polymer sequence: protein, RNA, or DNA. Query sequences must be at least 25 residues long and use the appropriate alphabet:
- Protein: ARNDCEQGHILKMFPSTWYVOUBZX
- RNA: ACUGNI
- DNA: ACTGUNI
This search uses MMseqs2 software (Steinegger and Söding, 2017) to identify sequences similar to your query. You can set a sequence identity cutoff to control how closely sequences must match. During the search, each candidate sequence is aligned to your query, and the number of identical residues is calculated. Sequences exceeding the cutoff are returned as search results.
Documentation
You can access the Sequence Similarity search by opening Advanced Search and clicking on (+) Sequence Similarity from the list of available search tools, or go directly to the search using this link: Sequence Similarity Search.
How to Provide a Query
The Sequence Similarity search supports two input modes
Enter a sequence
Paste or type a polymer sequence (protein, RNA, or DNA) directly into the input field. The sequence must be at least 25 residues long and use the correct alphabet for the polymer type.
Select an existing PDB or CSM sequence
Choose a polymer sequence from an existing PDB entry or Computed Structure Model (CSM) as your query. This allows you to quickly use a known structure as the basis for your search.
Search Options
The Sequence Similarity search lets you customize the following parameters to refine your results:
E-value
Filters results by statistical significance. Lower E-values indicate more significant matches. Only hits with E-values ≤ the selected cutoff will be returned.
Sequence Identity
Filters results based on how closely sequences match your query. Only sequences with identity ≥ the selected value are returned. Values range from 0 to 100.
Limitations of Sequence Similarity Search
Sequence similarity searches with short query sequences can match many sequences and provide false evolutionary relationships. This should be carefully examined and considered when making any conclusions about the matched sequences. For short sequences, the Sequence Motif search may be more suitable.
Query By Example
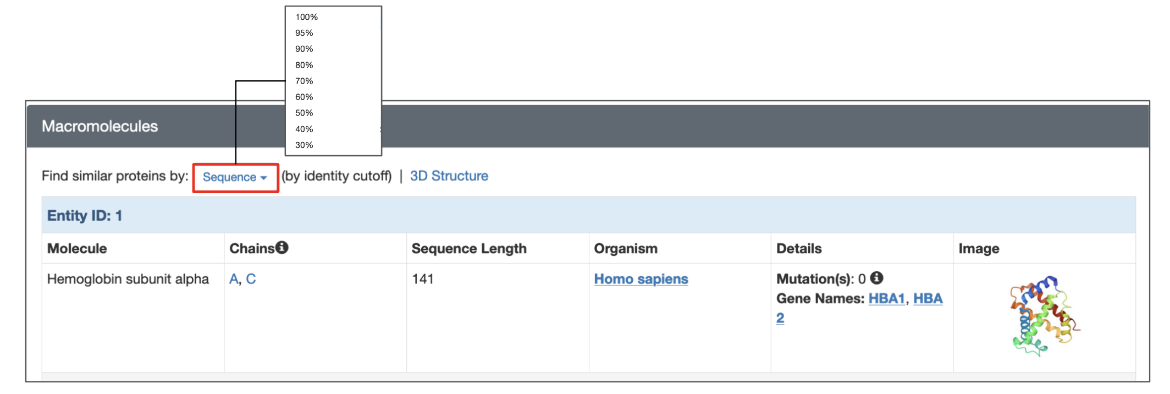
Each structure on RCSB.org has a dedicated Structure Summary page that displays information about the sequences in that entry in Macromolecules section. To search for polymer entities with sequences similar to a specific polymer entity, click the Sequence link above the entity details and select the desired sequence identity from the drop-down menu. The search is performed using the default E-value cutoff of 0.1.
|
| Options for launching a sequence similarity search from the structure summary page of a specific 3D structure. |
Search Results
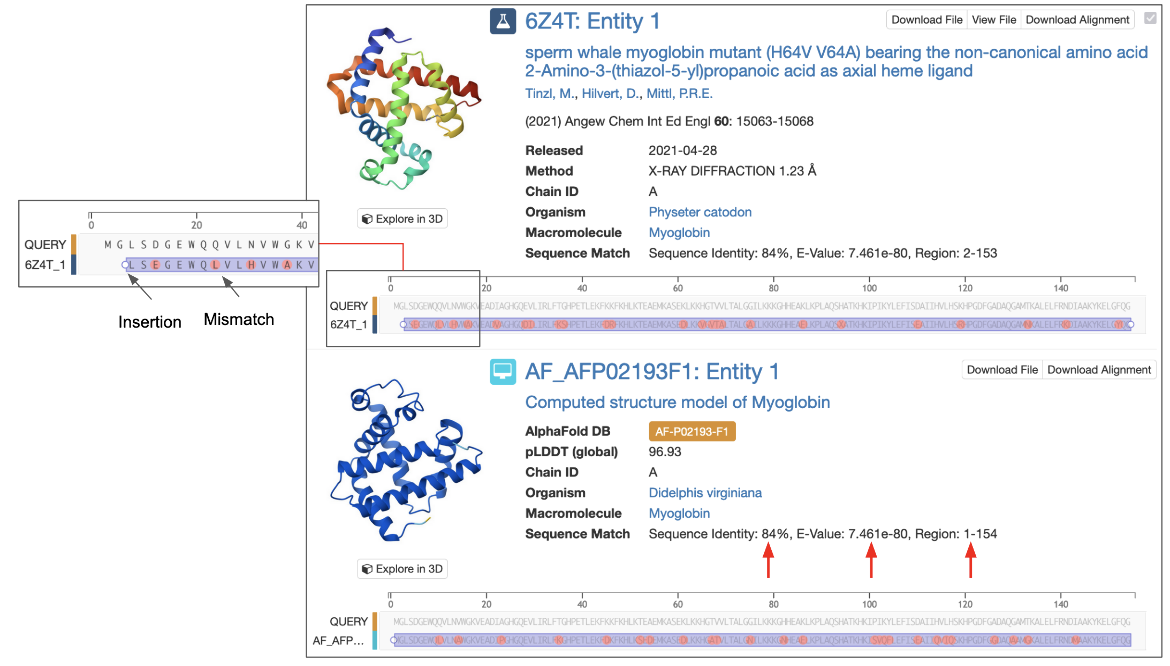
When a Sequence Similarity search is performed, results are displayed by default as matching Macromolecules. Each result shows alignment details, including sequence identity, E-value, and the aligned region (for example: Sequence Identity: 100%, E-value: 0, Region: 1–560):
- Sequence Identity is the ratio of the number of identical residues between the 2 aligned sequences over the aligned length, expressed as a percentage.
- E-Value provides a measure for whether the observed sequence identity was a chance match or if it has any evolutionary significance. This is reported as a number close to zero. The lower the E-Value, or the closer it is to zero, the more "significant" the match is. An E-Value of a significant match is often expressed in scientific notation, where the higher the exponent’s power the more significant the match (i.e., 1e-80 is a more significant match than 1e-30). Note that short query sequences may be seen in many other protein sequences by chance so these matches may have a high E-Value.
- Region specifies the residue ranges that were considered in the sequence comparison.
Alignments are presented using an interactive sequence viewer, allowing you to explore matches in detail. Zoom into the sequence and move the sequence left or right to examine sequence identities, mismatches, insertions, deletions or missing residues, etc. The Alignment Reference option lets you adjust the display to show the alignment from the perspective of the query, the subject, or a pairwise view. Learn more about these options and the color scheme used for alignment here.
Examples
Find sequences that are highly similar to the beta-lactamase in a PDB entry
- Use the PDB/CSM ID option to select the sequence from an existing PDB entry, such as pdb_00001blc.
- From the Select Sequence drop-down, choose beta-lactamase as your query sequence.
- Set the Sequence Identity cutoff to 90 to return polymer entity sequences sharing at least 90% identity with the query sequence.
- Click the Search button to return matching polymer entities
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of molecular biology, 215(3), 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Steinegger, M., Söding, J. (2017). MMseqs2 enables sensitive protein sequence similarity searching for the analysis of massive data sets. Nat Biotechnol 35, 1026–1028. https://doi.org/10.1038/nbt.3988