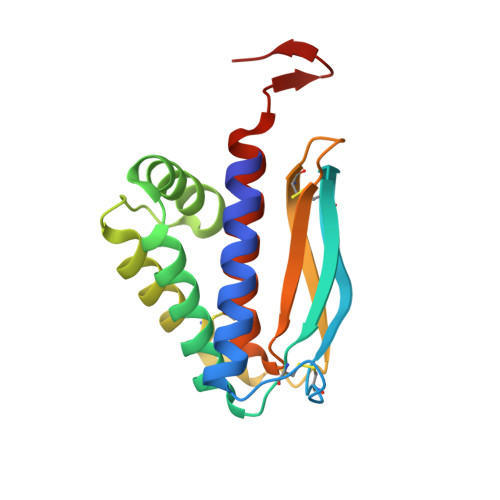
The 1.51-Angstrom structure of the poxvirus L1 protein, a target of potent neutralizing antibodies.
Su, H.P., Garman, S.C., Allison, T.J., Fogg, C., Moss, B., Garboczi, D.N.(2005) Proc Natl Acad Sci U S A 102: 4240-4245
- PubMed: 15761054
- DOI: https://doi.org/10.1073/pnas.0501103102
- Primary Citation of Related Structures:
1YPY - PubMed Abstract:
Although eradicated from nature more than two decades ago, the threat of smallpox has reemerged because of concerns over its use as a biological weapon. We present the structure of the poxvirus L1 protein, a molecule that is conserved throughout the poxvirus family and is nearly identical in vaccinia virus and in variola virus, which causes smallpox. L1 is a myristoylated envelope protein that is a potent target for neutralizing antibodies and an important component of current experimental vaccines. The L1 structure reveals a hydrophobic cavity located adjacent to its N terminus. The cavity would be capable of shielding the myristate moiety, which is essential for virion assembly. The structure of L1 is a step in the elucidation of molecular mechanisms common to all poxviruses that may stimulate the design of safer vaccines and new antipoxvirus drugs.
- Structural Biology Section, Laboratory of Immunogenetics, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 12441 Parklawn Drive, Rockville, MD 20852, USA.
Organizational Affiliation: