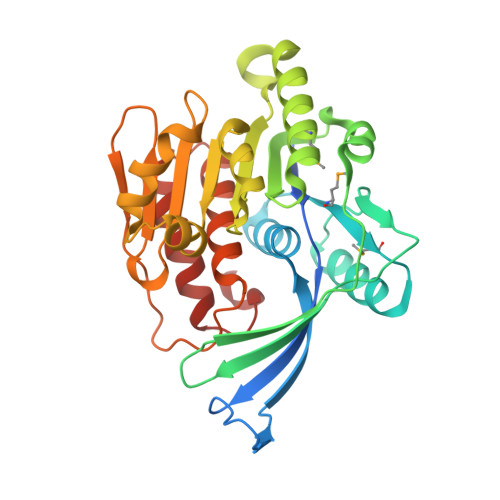
Residues in the fructose-binding pocket are required for ketohexokinase-A activity.
Ferreira, J.C., Villanueva, A.J., Fadl, S., Al Adem, K., Cinviz, Z.N., Nedyalkova, L., Cardoso, T.H.S., Andrade, M.E., Saksena, N.K., Sensoy, O., Rabeh, W.M.(2024) J Biological Chem 300: 107538-107538
- PubMed: 38971308
- DOI: https://doi.org/10.1016/j.jbc.2024.107538
- Primary Citation of Related Structures:
2HLZ - PubMed Abstract:
Excessive fructose consumption is a primary contributor to the global surges in obesity, cancer, and metabolic syndrome. Fructolysis is not robustly regulated and is initiated by ketohexokinase (KHK). In this study, we determined the crystal structure of KHK-A, one of two human isozymes of KHK, in the apo-state at 1.85 Å resolution, and we investigated the roles of residues in the fructose-binding pocket by mutational analysis. Introducing alanine at D15, N42, or N45 inactivated KHK-A, whereas mutating R141 or K174 reduced activity and thermodynamic stability. Kinetic studies revealed that the R141A and K174A mutations reduced fructose affinity by 2- to 4-fold compared to WT KHK-A, without affecting ATP affinity. Molecular dynamics simulations provided mechanistic insights into the potential roles of the mutated residues in ligand coordination and the maintenance of an open state in one monomer and a closed state in the other. Protein-protein interactome analysis indicated distinct expression patterns and downregulation of partner proteins in different tumor tissues, warranting a reevaluation of KHK's role in cancer development and progression. The connections between different cancer genes and the KHK signaling pathway suggest that KHK is a potential target for preventing cancer metastasis. This study enhances our understanding of KHK-A's structure and function and offers valuable insights into potential targets for developing treatments for obesity, cancer, and metabolic syndrome.
- Science Division, New York University Abu Dhabi, Abu Dhabi, United Arab Emirates.
Organizational Affiliation: