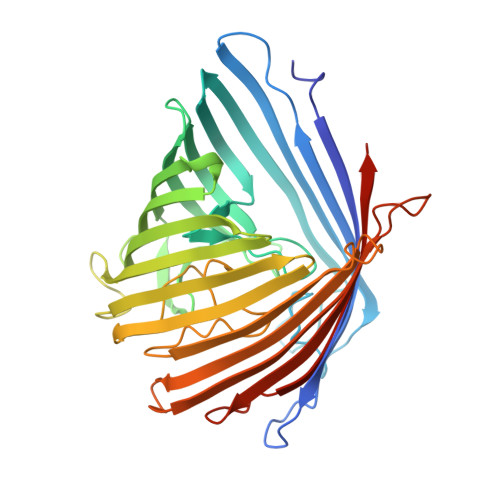
Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution.
Sampathkumar, P., Lu, F., Zhao, X., Li, Z., Gilmore, J., Bain, K., Rutter, M.E., Gheyi, T., Schwinn, K.D., Bonanno, J.B., Pieper, U., Fajardo, J.E., Fiser, A., Almo, S.C., Swaminathan, S., Chance, M.R., Baker, D., Atwell, S., Thompson, D.A., Emtage, J.S., Wasserman, S.R., Sali, A., Sauder, J.M., Burley, S.K.(2010) Proteins 78: 3056-3062
- PubMed: 20737437
- DOI: https://doi.org/10.1002/prot.22829
- Primary Citation of Related Structures:
3JTY - PubMed Abstract:
The X-ray structure of a putative BenF-like (gene name: PFL1329) protein from Pseudomonas fluorescens Pf-5 (PflBenF) has been determined at 2.6Å resolution. X-ray crystallography revealed a canonical 18-stranded β-barrel fold that forms a central pore with a diameter of ∼4.6Å, which is consistent with the size and physicochemical properties of the presumed aromatic acid substrate, benzoate. Detailed comparisons with the previously-determined structure of Pseudomonas aeruginosa OpdK, a vanillate influx channel, revealed an arginine-rich aromatic acid selectivity filter of nearly identical structure composed of seven highly conserved residues Arg∼Asp∼Arg∼Arg∼Ser∼Asp∼Arg (R∼D∼R∼R∼S∼D∼R sequence motif, where ∼ denotes intervening residues) that define the narrowest part of the pore.
- New York SGX Research Center for Structural Genomics, Eli Lilly and Company, Lilly Biotechnology Center, San Diego, California 92121, USA. sampathkumarpa@lilly.com
Organizational Affiliation: