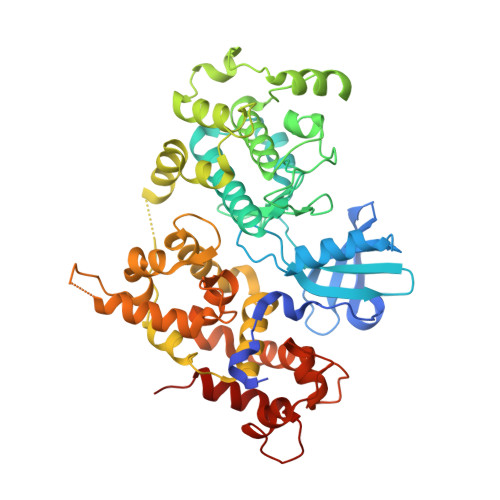
Crystal structure of full length CpCDPK3 (cgd5_820) in complex with Ca2+ and AMPPNP
Qiu, W., Hutchinson, A., Wernimont, A., Walker, J.R., Sullivan, H., Lin, Y.-H., Mackenzie, F., Kozieradzki, I., Cossar, D., Schapira, M., Senisterra, G., Vedadi, M., Arrowsmith, C.H., Bountra, C., Weigelt, J., Edwards, A.M., Bochkarev, A., Hui, R., Amani, M.To be published.