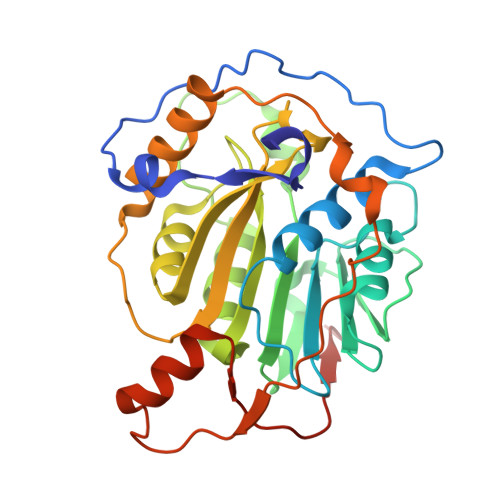
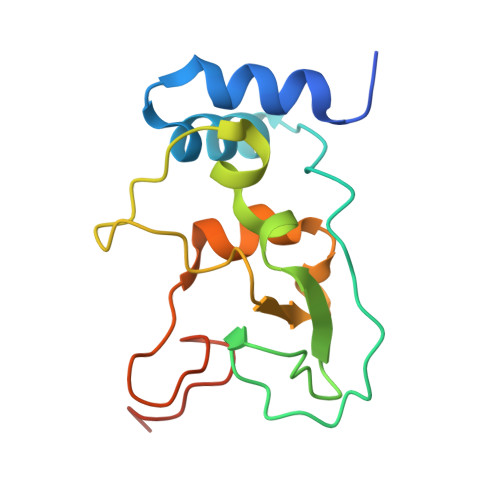
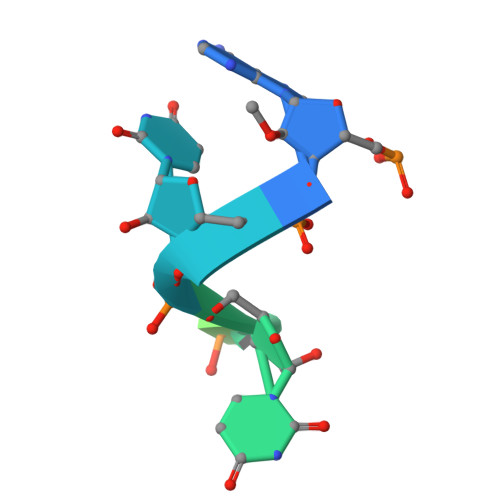
Structural insights into the assembly and regulation of 2'-O RNA methylation by SARS-CoV-2 nsp16/nsp10.
Misra, A., Rahisuddin, R., Parihar, M., Arya, S., Viswanathan, T., Jackson, N., Qi, S., Chan, S.H., Harris, R.S., Martinez-Sobrido, L., Gupta, Y.K.(2025) Structure 33: 1027-1039.e4
- PubMed: 40220753
- DOI: https://doi.org/10.1016/j.str.2025.03.009
- Primary Citation of Related Structures:
8VUO - PubMed Abstract:
2'-O-ribose methylation of the first transcribed base (adenine or A 1 in SARS-CoV-2) of viral RNA mimics host RNAs and subverts the innate immune response. How nsp16, with partner nsp10, assembles on the 5'-end of SARS-CoV-2 mRNA to methylate A 1 is not fully understood. We present a ∼2.4 Å crystal structure of the heterotetrameric complex formed by the cooperative assembly of two nsp16/nsp10 heterodimers with one 10-mer Cap-1 RNA (product) bound to each. An aromatic zipper-like motif in nsp16 and the N-terminal regions of nsp10 and nsp16 orchestrate oligomeric assembly for efficient methylation. The front catalytic pocket of nsp16 stabilizes the upstream portion of the RNA while downstream RNA remains unresolved, likely due to flexibility. An inverted nsp16 dimer extends the positively charged surface for longer RNA to influence catalysis. Additionally, a non-specific nucleotide-binding pocket on the backside of nsp16 plays a critical role in catalysis, contributing to enzymatic activity.
- Greehey Children's Cancer Research Institute, University of Texas Health Science Center, San Antonio, TX 78229, USA; Department of Biochemistry and Structural Biology, University of Texas Health Science Center, San Antonio, TX 78229, USA.
Organizational Affiliation: