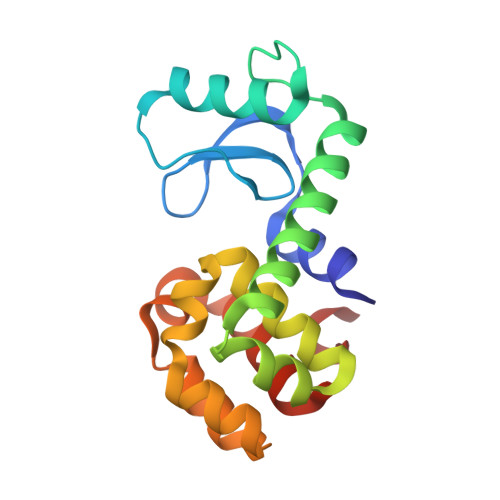
Structural characterization of the Cu(II)-NTA spin label on alpha-helices by X-ray crystallography and electron paramagnetic resonance
Besaw, J.E., Reichenwallner, J., Chen, E.Y., Hermet-Teesalu, P., Tregubenko, A., Kim, K., Morizumi, T., Ustav Jr., M., Ernst, O.P.(2026) Structure 34