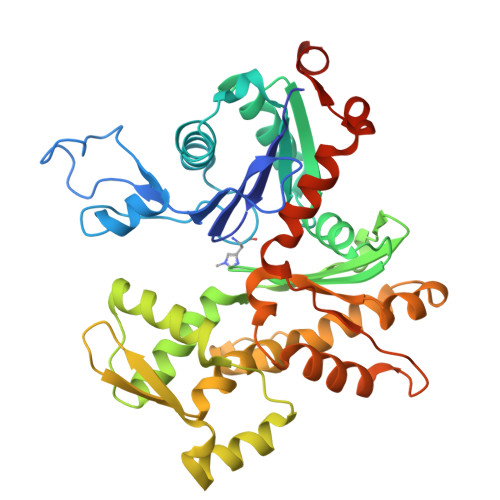
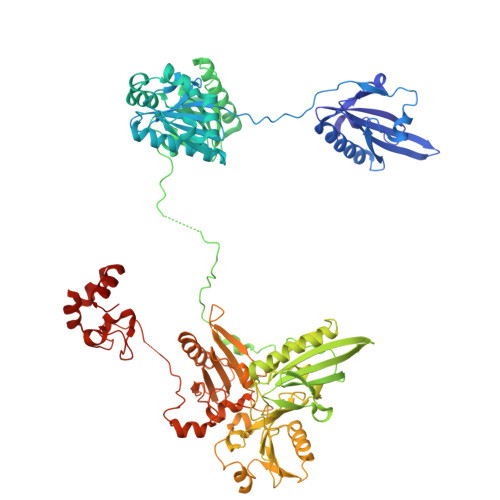
The structure of an actin nucleus stabilized by villin.
Robinson, R.C., Chongrungreang, T., Ponlachantra, K., Boonyarit, B., Dilly, G.F., Li, Y.I., Girguis, P.R., Copley, R.R., Claridge-Chang, A.(2025) Sci Adv 11: eadw6915-eadw6915
- PubMed: 41337577
- DOI: https://doi.org/10.1126/sciadv.adw6915
- Primary Citation of Related Structures:
9JUS, 9JVT, 9JW0 - PubMed Abstract:
Villin is an actin filament nucleating, severing, capping and bundling protein; however, the structural basis for villin's functions and the characteristics of the actin polymerization nucleus remain poorly understood. Here, we present the structure of vent-worm villin bound to a trimeric actin nucleus. Villin wraps around and caps the barbed end of the actin trimer. Its headpiece domain interacts at the junction of two laterally associated actin protomers, leaving the pointed-end subunits open for elongation. Within the actin trimer, the two longitudinally associated subunits adopt barbed and pointed-end subunit conformations, while the lateral protomer exhibits a monomeric conformation. This provides the first view of an actin-filament nucleus, revealing that the transition into the filamentous form is stimulated and stabilized by the interactions with the pointed-end subunits. Our results also illuminate mechanisms of actin-filament dynamics and villin capping and severing, suggesting that F-to-G actin conformational transitions facilitate the later process.
- School of Biomolecular Science and Engineering (BSE), Vidyasirimedhi Institute of Science and Technology (VISTEC), Rayong 21210 Thailand.
Organizational Affiliation: