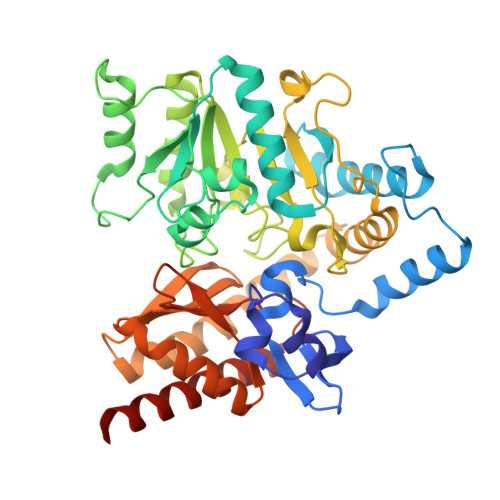
Biochemical characterization and mutational analysis of the tetrameric DABA transaminase EctB from the Arctic bacterium Marinobacter sp. CK1.
Skogvold, A.C.A., Leiros, I., Engh, R.A., Erlandsen, H.(2026) FEBS J
- PubMed: 41652856
- DOI: https://doi.org/10.1111/febs.70441
- Primary Citation of Related Structures:
9S1P, 9S1Q - PubMed Abstract:
Ectoine is a chemical chaperone and osmoprotectant discovered in halophilic bacteria where it is used to protect from osmotic stress in high-salt environments. Ectoine has become a valuable product in pharmaceutical, biotechnological, cosmetical, and other applications due to its protective and stabilizing properties. The rate-limiting step of ectoine biosynthesis is catalyzed by the l-2,4-diaminobutyric acid (DABA) transaminase enzyme EctB, which converts aspartate-β-semialdehyde (ASA) to DABA. We studied the structural and functional properties of EctB from a novel halophilic bacterium Marinobacter sp. CK1. Crystal structures of complexes with mEctB/pyridoxamine-5'-phosphate and an inactive mutant K264A/pyridoxal 5'-phosphate reveal that the enzyme forms tetramers. We also investigated the stability/activity relationship and validated residues important for activity and flexibility by characterizing mEctB and a series of 17 mutants for oligomerization, thermal stability, and catalytic activity. The wild-type enzyme retained > 50% activity over a pH range of 2.5 units, with an optimum at pH 8, all NaCl concentrations (0-1 m), and temperatures between 30 °C and 45 °C. Enzyme activity was highest using DABA and GABA as substrates. Mutations at the dimer-dimer interface (K150A, E194A, E194Q, and N134A) overall lowered the enzyme's thermal stability while two (E194Q and N134A) completely abolished activity. Mutation of Arg96 at the surface significantly lowered activity and melting temperature, but with little effect on oligomerization. The active site mutants K264, Y14 and R295 were shown to be vital for activity. Overall, this study provides new information regarding the structure, stability and function of EctB.
- Department of Chemistry, Faculty of Science and Technology, The Norwegian Structural Biology Center, UiT the Arctic University of Norway, Tromsø, Norway.
Organizational Affiliation: