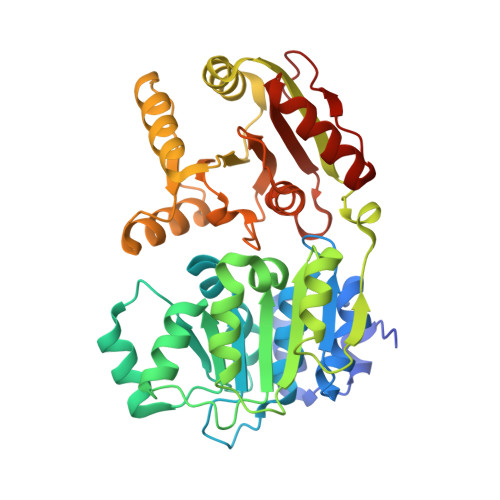
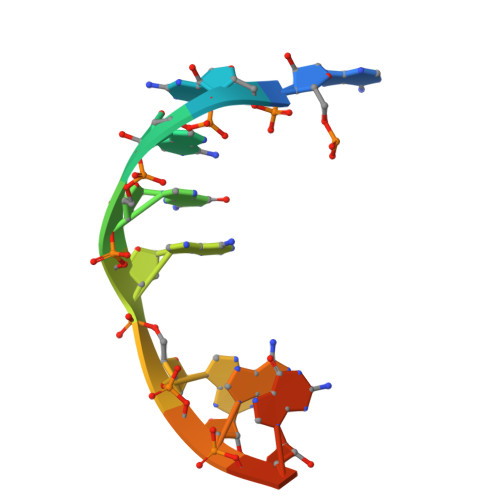
Structural Basis for the Improved RNA Clamping of Amidino-Rocaglates to eIF4A1.
Conley, J.F., Brown, L.E., McNeely, J.H., Pelletier, J., Porco Jr., J.A., Allen, K.N.(2025) ACS Omega 10: 5795-5808
- PubMed: 39989799
- DOI: https://doi.org/10.1021/acsomega.4c09421
- Primary Citation of Related Structures:
9DTS - PubMed Abstract:
Eukaryotic initiation factor 4A-1 (eIF4A1) is an ATP-dependent RNA helicase that unwinds 5'-UTR mRNA secondary structures to facilitate cap-dependent translation initiation. Rocaglates, a class of natural products typified by rocaglamide A (RocA), possess antineoplastic and anti-infectious activity mediated by their interaction with eIF4A1. Rocaglates inhibit cap-dependent translation initiation by "clamping" eIF4A1 onto polypurine RNA, which impedes ribosome scanning. A novel class of rocaglate derivatives, amidino-rocaglates (ADRs) which feature an amidine ring fused to the rocaglate core, is particularly effective at promoting eIF4A1-RNA-clamping compared to other rocaglate congeners. Herein, we present the X-ray crystal structure of an ADR in complex with eIF4A1, the nonhydrolyzable ATP ground-state mimic adenylyl-imidodiphosphate (AMPPNP), and poly r(AG) 5 RNA refined to 1.69 Å resolution. The binding pose and interactions of the ADR with eIF4A1 do not differ substantially from those of RocA, prompting an investigation of the basis for enhanced target engagement. Computational modeling suggests that the rigidified ADR scaffold is inherently preorganized in an eIF4A1-RNA binding-competent conformation, thereby avoiding entropic penalties associated with RocA binding. This study illustrates how conformational rigidification of the rocaglate scaffold can be leveraged to improve potency for the development of rocaglates as potential anticancer and anti-infectious agents.
- Department of Pharmacology, Physiology & Biophysics, Boston University, Boston, Massachusetts 02215, United States.
Organizational Affiliation: