Tandem antigen chimera of Pfs230 and Pfs48/45 bound by potent mAbs
Ivanochko, D., Semesi, A., Julien, J.P.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RUPA-39 heavy chain | 227 | Homo sapiens | Mutation(s): 0 | 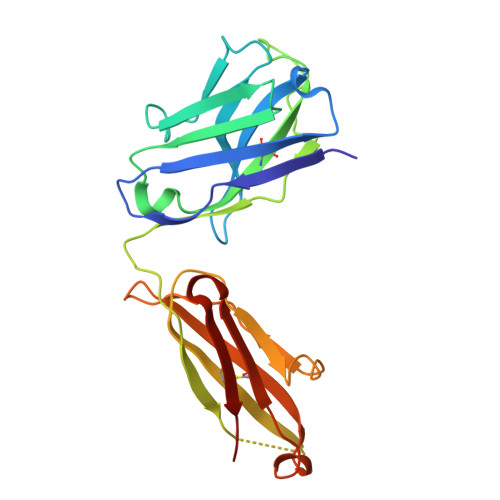 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RUPA-39 Kappa chain | 215 | Homo sapiens | Mutation(s): 0 | 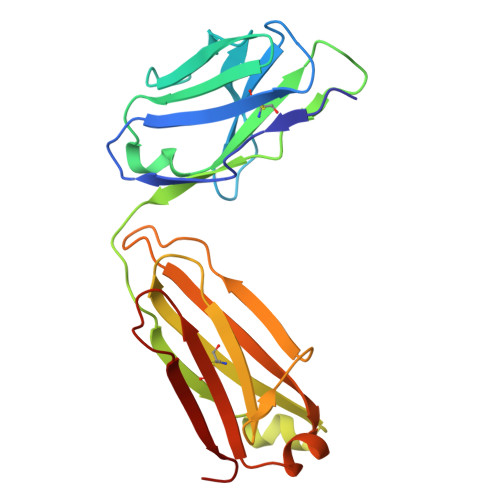 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RUPA-44 Kappa chain | C [auth E] | 214 | Homo sapiens | Mutation(s): 0 | 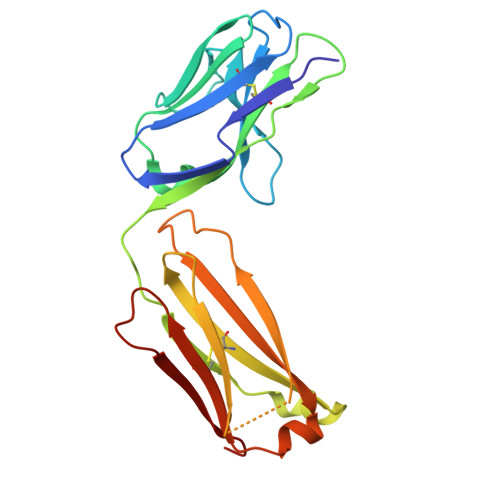 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RUPA-44 heavy chain | D [auth F] | 230 | Homo sapiens | Mutation(s): 0 | 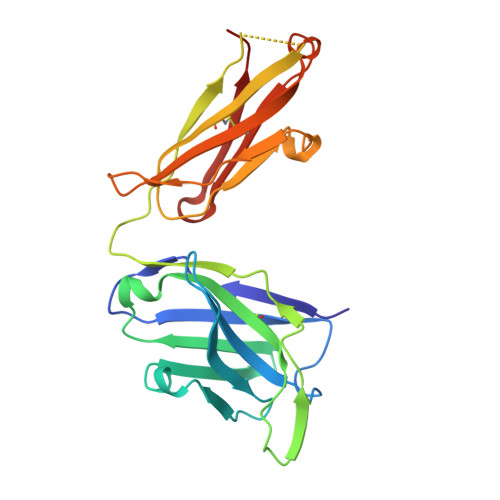 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gametocyte surface protein P230,Gametocyte surface protein P45/48 | E [auth I] | 324 | Plasmodium falciparum 3D7 | Mutation(s): 0 Gene Names: PFS230, PF230, S230, PF3D7_0209000, PF45/48, PFS45-48, PFS45/48, PF13_0247, PF3D7_1346700 |  |
UniProt | |||||
Find proteins for P68874 (Plasmodium falciparum (isolate 3D7)) Explore P68874 Go to UniProtKB: P68874 | |||||
Find proteins for Q8I6T1 (Plasmodium falciparum (isolate 3D7)) Explore Q8I6T1 Go to UniProtKB: Q8I6T1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | P68874Q8I6T1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 8 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PGE Query on PGE | J [auth A] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| CAC Query on CAC | JA [auth F], LA [auth F] | CACODYLATE ION C2 H6 As O2 OGGXGZAMXPVRFZ-UHFFFAOYSA-M |  | ||
| TRS Query on TRS | S [auth B] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| PEG Query on PEG | N [auth B], P [auth B], T [auth B], U [auth B] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | BA [auth E] CA [auth E] DA [auth E] EA [auth E] G [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | AA [auth E], FA [auth E], GA [auth E], IA [auth F], V [auth B] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| CA Query on CA | HA [auth E], W [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | K [auth A], X [auth B], Y [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 227.99 | α = 90 |
| b = 80.49 | β = 111.46 |
| c = 137.73 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| STARANISO | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Bill & Melinda Gates Foundation | United States | INV-008866 and OPP1156262 |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | 1R01AI148557-01A1 |
| Canadian Institutes of Health Research (CIHR) | Canada | 428410 |