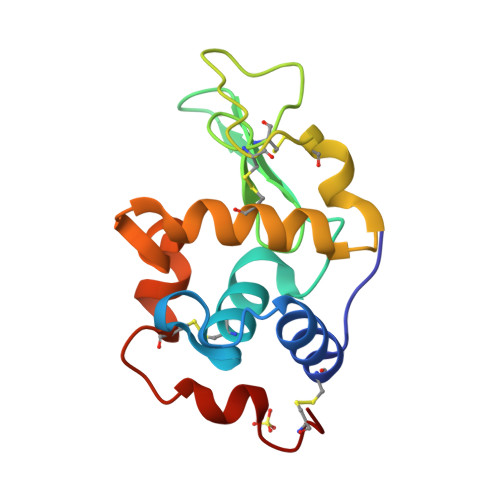
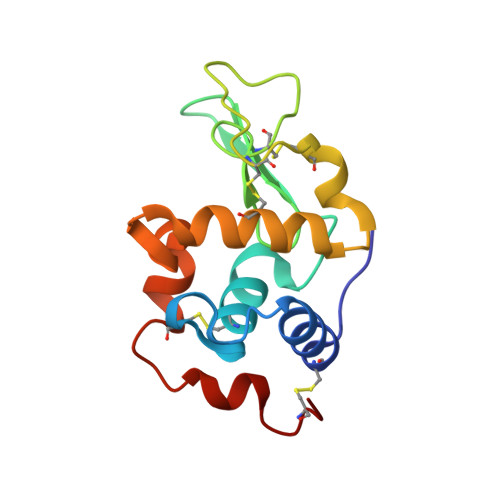
The 1.9 A X-ray structure of egg-white lysozyme from Taiwanese soft-shelled turtle (Trionyx Sinensis Wiegmann) exhibits structural differences from the standard chicken-type lysozyme.
Siritapetawee, J., Thammasirirak, S., Robinson, R.C., Yuvaniyama, J.(2009) J Biochem 145: 193-198
- PubMed: 19029145
- DOI: https://doi.org/10.1093/jb/mvn156
- Primary Citation of Related Structures:
2GV0 - PubMed Abstract:
Lysozyme from Taiwanese soft-shelled turtle (SSTLB) has been purified from turtle egg white and crystallized. The crystals diffract X-rays beyond 2 A resolution and belong to the orthorhombic P2(1)2(1)2(1) space group containing one molecule per asymmetric unit. The structure was elucidated using pheasant egg-white lysozyme as the molecular replacement search template. The overall structure of SSTLB is very similar to that of hen egg-white lysozyme (HEWL). Nevertheless, Pro104 in the substrate subsite A and other amino acids in the substrate subsites E and F differ from those of HEWL. These substitutions result in local conformational differences in the substrate binding sites of the two enzymes, effecting substrate binding and transglycosylation efficiency, translating into differing ranges of products.
Organizational Affiliation:
College of Medicine and Public Health, Ubon Rajathanee University, Ubon Ratchathani, Thailand.