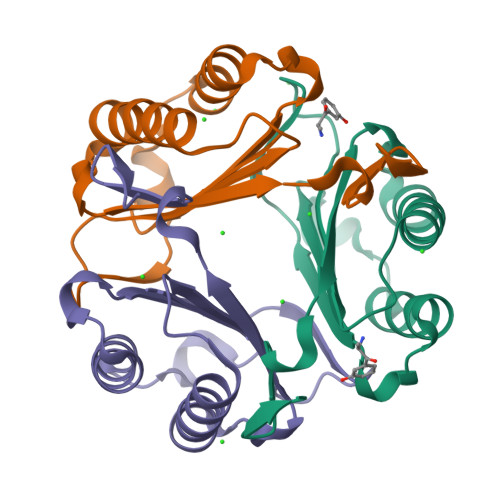
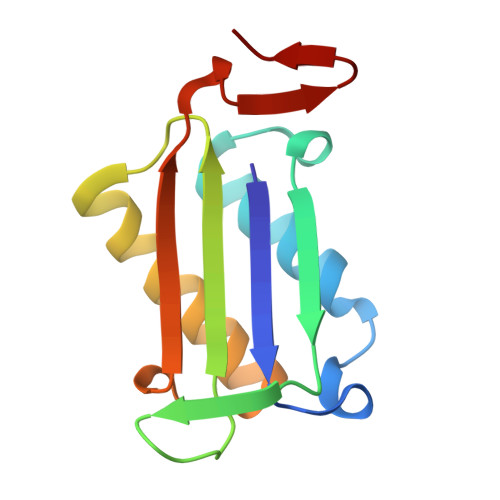
Repurposing old drugs as novel inhibitors of human MIF from structural and functional analysis.
Yang, L., Yang, C., Wang, L., Yang, Z., Guo, D., Fan, C.(2022) Bioorg Med Chem Lett 55: 128445-128445
- PubMed: 34758374
- DOI: https://doi.org/10.1016/j.bmcl.2021.128445
- Primary Citation of Related Structures:
7EDQ, 7EE8 - PubMed Abstract:
Human macrophage migration inhibitory factor (MIF) is an important pro-inflammatory cytokine that plays multiple pleiotropic functions. It is considered as a promising therapeutic target for the infectious, autoimmune, and cardiovascular diseases and cancers. The development of MIF inhibitors has not been translated into clinical success despite decades of research. Given the time and cost of developing new drugs, existing drugs with clarified safety and pharmacokinetics are explored for their potential as novel MIF inhibitors. This study identified five known drugs that could inhibit MIF's tautomerase activity and MIF-mediated cell chemotaxis in RAW264.7 cells. It was found that compounds D2 (histamine), D5 (metaraminol), and D8 (nebivolol) exhibited micromolar-range inhibition potency close to the positive control ISO-1. Kinetics and the mechanism for inhibition were subsequently determined. Moreover, the detailed inhibitor-binding patterns were investigated by X-ray crystallography, computational molecular docking, and structure-based analysis. Therefore, this study elucidates the molecular mechanism of repurposed drugs acting on MIF and provides a structural foundation for lead optimization to promote the clinical development of MIF-targeted drugs.
Organizational Affiliation:
School of Basic Medical Sciences, Wuhan University, Wuhan 430071, PR China.