Crystal structure of the liprin-alpha2/RIM1 complex
Jin, G., Wei, Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Regulating synaptic membrane exocytosis 1 | 175 | Rattus norvegicus | Mutation(s): 0 Gene Names: Rims1 | 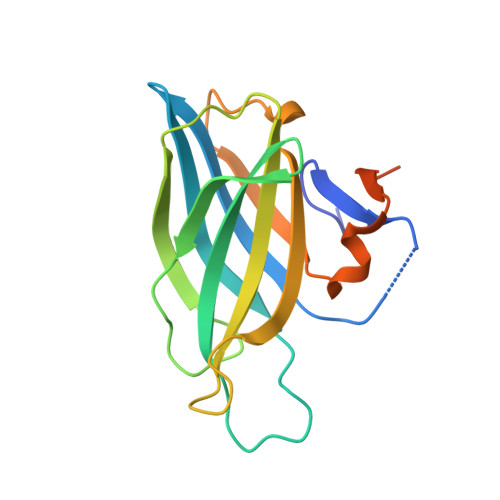 | |
UniProt | |||||
Find proteins for A0A8I6GEN0 (Rattus norvegicus) Explore A0A8I6GEN0 Go to UniProtKB: A0A8I6GEN0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8I6GEN0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Liprin-alpha-2 | 111 | Homo sapiens | Mutation(s): 0 Gene Names: PPFIA2 | 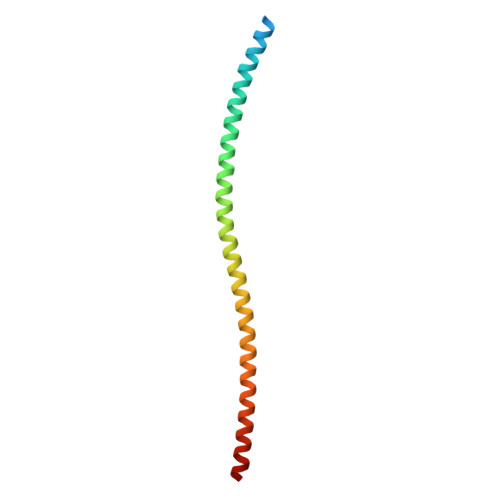 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O75334 (Homo sapiens) Explore O75334 Go to UniProtKB: O75334 | |||||
PHAROS: O75334 GTEx: ENSG00000139220 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O75334 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.663 | α = 90 |
| b = 92.697 | β = 102.95 |
| c = 66.583 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| SCALEPACK | data scaling |
| HKL-2000 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | China | RCJC20210609104333007 |