Cryo-EM structure of GPCR16-Gi2 complex
Wang, X., Zhou, C., Wu, L.J., Hua, T., Liu, Z.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(i) subunit alpha-2 | 355 | Homo sapiens | Mutation(s): 3 Gene Names: GNAI2, GNAI2B | 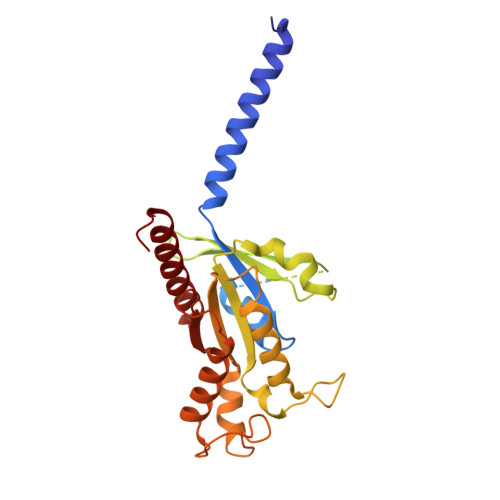 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04899 (Homo sapiens) Explore P04899 Go to UniProtKB: P04899 | |||||
PHAROS: P04899 GTEx: ENSG00000114353 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04899 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | 366 | Homo sapiens | Mutation(s): 0 Gene Names: GNB1 | 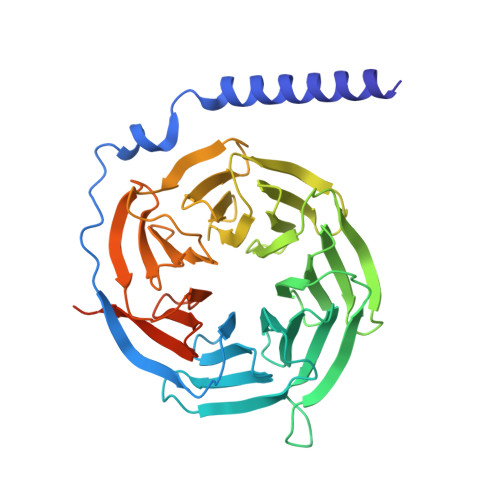 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62873 (Homo sapiens) Explore P62873 Go to UniProtKB: P62873 | |||||
PHAROS: P62873 GTEx: ENSG00000078369 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62873 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | C [auth G] | 71 | Homo sapiens | Mutation(s): 0 Gene Names: GNG2 | 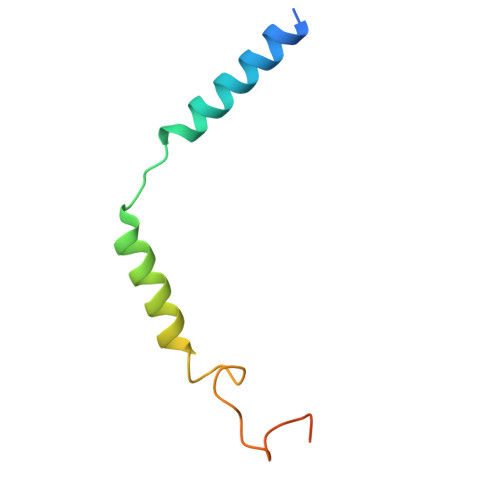 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P59768 (Homo sapiens) Explore P59768 Go to UniProtKB: P59768 | |||||
PHAROS: P59768 GTEx: ENSG00000186469 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| scFv16 | D [auth S] | 266 | Homo sapiens | Mutation(s): 0 | 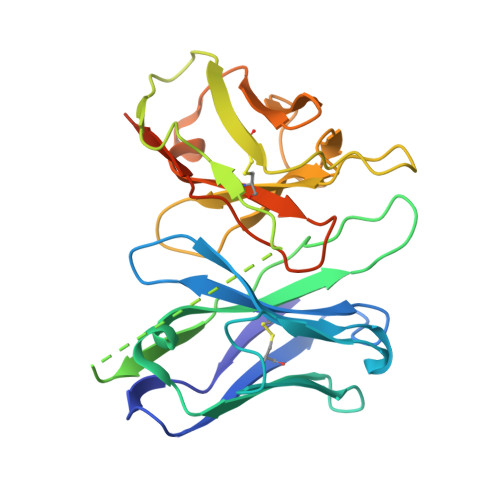 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| exo-alpha-sialidase,Taste receptor type 2 member 16,LgBiT | E [auth R] | 1,011 | Streptococcus pneumoniae, Homo sapiens, synthetic construct This entity is chimeric | Mutation(s): 1 Gene Names: nanA_2, SAMEA3353537_00427, TAS2R16 EC: 3.2.1.18 | 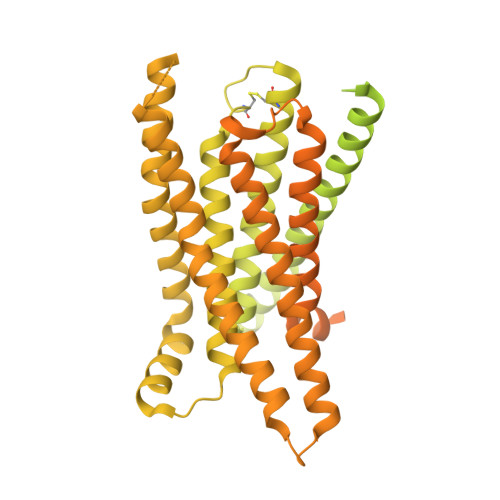 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9NYV7 (Homo sapiens) Explore Q9NYV7 Go to UniProtKB: Q9NYV7 | |||||
PHAROS: Q9NYV7 GTEx: ENSG00000128519 | |||||
Find proteins for A0A4J2AMT3 (Streptococcus pneumoniae) Explore A0A4J2AMT3 Go to UniProtKB: A0A4J2AMT3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q9NYV7A0A4J2AMT3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SA0 (Subject of Investigation/LOI) Query on SA0 | F [auth R] | 2-(hydroxymethyl)phenyl beta-D-glucopyranoside C13 H18 O7 NGFMICBWJRZIBI-UJPOAAIJSA-N |  | ||