Structure of western equine encephalitis virus CBA87 VLP in complex with human PCDH10 EC1
Abraham, J., Fan, X., Li, W.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Structural polyprotein | A [auth D], D [auth G], G [auth J], M [auth A] | 434 | Western equine encephalitis virus | Mutation(s): 0 | 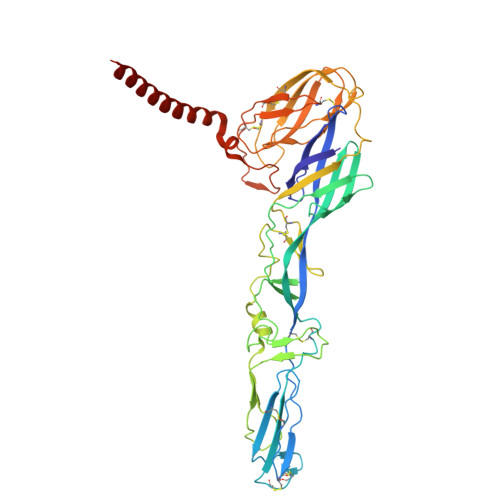 |
UniProt | |||||
Find proteins for C7EPF2 (Western equine encephalitis virus) Explore C7EPF2 Go to UniProtKB: C7EPF2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C7EPF2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Structural polyprotein | B [auth E], E [auth H], H [auth K], N [auth B] | 403 | Western equine encephalitis virus | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q1W679 (Western equine encephalitis virus) Explore Q1W679 Go to UniProtKB: Q1W679 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q1W679 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Capsid protein | C [auth F], F [auth I], I [auth L], O [auth C] | 163 | Western equine encephalitis virus | Mutation(s): 0 EC: 3.4.21.90 |  |
UniProt | |||||
Find proteins for P13897 (Western equine encephalitis virus) Explore P13897 Go to UniProtKB: P13897 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13897 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protocadherin-10 | J [auth M], K [auth N], L [auth O], P | 103 | Homo sapiens | Mutation(s): 0 Gene Names: PCDH10, KIAA1400 | 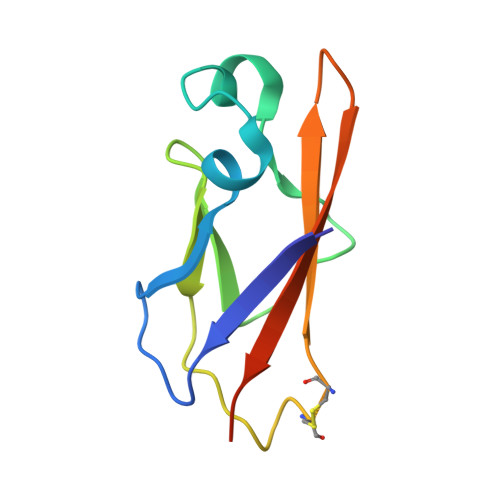 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9P2E7 (Homo sapiens) Explore Q9P2E7 Go to UniProtKB: Q9P2E7 | |||||
PHAROS: Q9P2E7 GTEx: ENSG00000138650 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9P2E7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | AA [auth K] BA [auth K] CA [auth A] DA [auth A] EA [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | -- |
| Howard Hughes Medical Institute (HHMI) | United States | -- |